The easiest way to install Snakemake (and other bioinformatics software) is via conda, which you get through Anaconda or miniconda.

wget https://repo.anaconda.com/archive/Anaconda3-5.3.1-Linux-x86_64.sh
bash Anaconda3-5.3.1-Linux-x86_64.sh
Or for miniconda run
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
bash Miniconda3-latest-Linux-x86_64.sh
Why Conda?
Conda has the advantages:
- Language agnostic
- “Easy” installation of software/environment by:
- One line installation
- Autoresolves dependencies
- Tracks versions
- Environment isolation:
- Install conflicting software into different environments
- Install different versions of software to different environments
There is a handy Cheat Sheet for conda commands.
Conda vs Containers
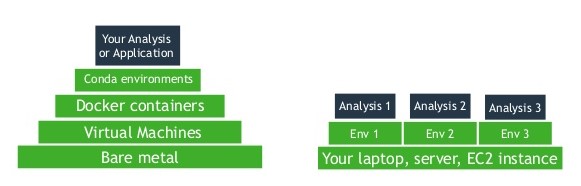
Conda:
- Is lighter weight than VM/Containers.
- Aggregates all the dependencies on top of existing OS.
- Does NOT isolate your environment from the underlying OS.
| VM | Containers | Conda | |
|---|---|---|---|
| Drivers | Included | System | System |
| OS | Included | Included | System |
| Programs | Included | Included | Included |
Important: I swiped this cool table from Jiangwei Yao, Ph.D. who gave a training through OAMD titled Building your own personal computing environment using Conda.